Foundation 1: Biomolecules covers essential biochemistry concepts, including the structure and function of proteins, carbohydrates, lipids, and nucleic acids. Understanding metabolic pathways, genetic information flow, and cellular regulation provides a solid basis for tackling the Biological and Biochemical Foundations section. Practice passages reinforce critical thinking and application, helping to excel on the MCAT’s challenging questions.
Learning Objectives
In studying “Foundation 1: Biomolecules” MCAT Biological and Biochemical Foundations of Living Systems, you should learn to identify the structure and function of key biomolecules, including proteins, carbohydrates, lipids, and nucleic acids. Analyze the role of these molecules in biological processes such as metabolism, energy storage, and genetic information transfer. Evaluate the biochemical pathways involved in cellular functions, including enzyme kinetics, thermodynamics, and molecular interactions. Additionally, practice interpreting experimental data, applying biochemical principles to biological systems, and integrating your knowledge to solve complex problems related to living organisms and their biochemical foundations.
1. Structure and Function of Biomolecules
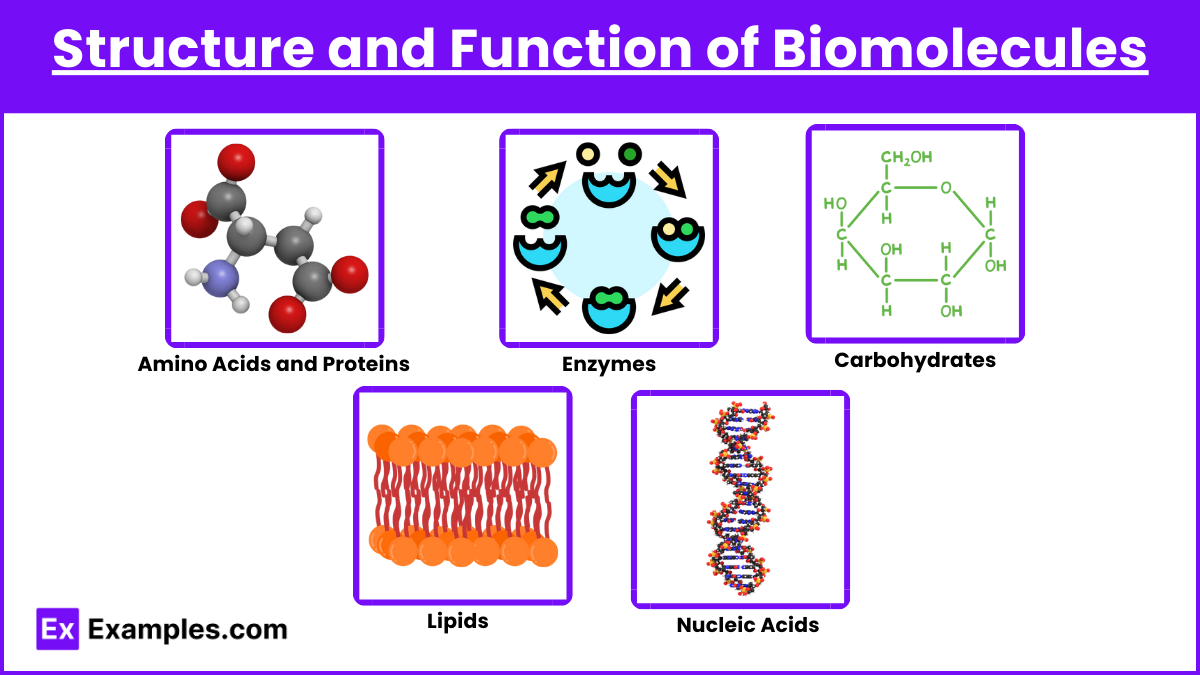
- Amino Acids and Proteins: Know the structure of amino acids, their classifications (e.g., hydrophobic, hydrophilic), and how they form peptide bonds. Understand protein structure (primary, secondary, tertiary, and quaternary) and the significance of each level to protein function.
- Enzymes: Focus on enzyme kinetics, including the Michaelis-Menten equation, concepts of Vmax and Km, and enzyme inhibition types (competitive, noncompetitive, and uncompetitive). Grasp the role of coenzymes, cofactors, and the impact of pH and temperature on enzyme activity.
- Carbohydrates: Understand monosaccharides, disaccharides, and polysaccharides, as well as glycosidic linkages and carbohydrate functions in energy storage (e.g., glycogen) and structure (e.g., cellulose).
- Lipids: Familiarize yourself with fatty acids, triglycerides, phospholipids, and cholesterol. Understand their roles in membrane structure, energy storage, and as signaling molecules.
- Nucleic Acids: Cover DNA and RNA structure, including base pairing and the double-helix model. Understand replication, transcription, translation, and the roles of mRNA, tRNA, and rRNA in protein synthesis.
2. Metabolic Pathways
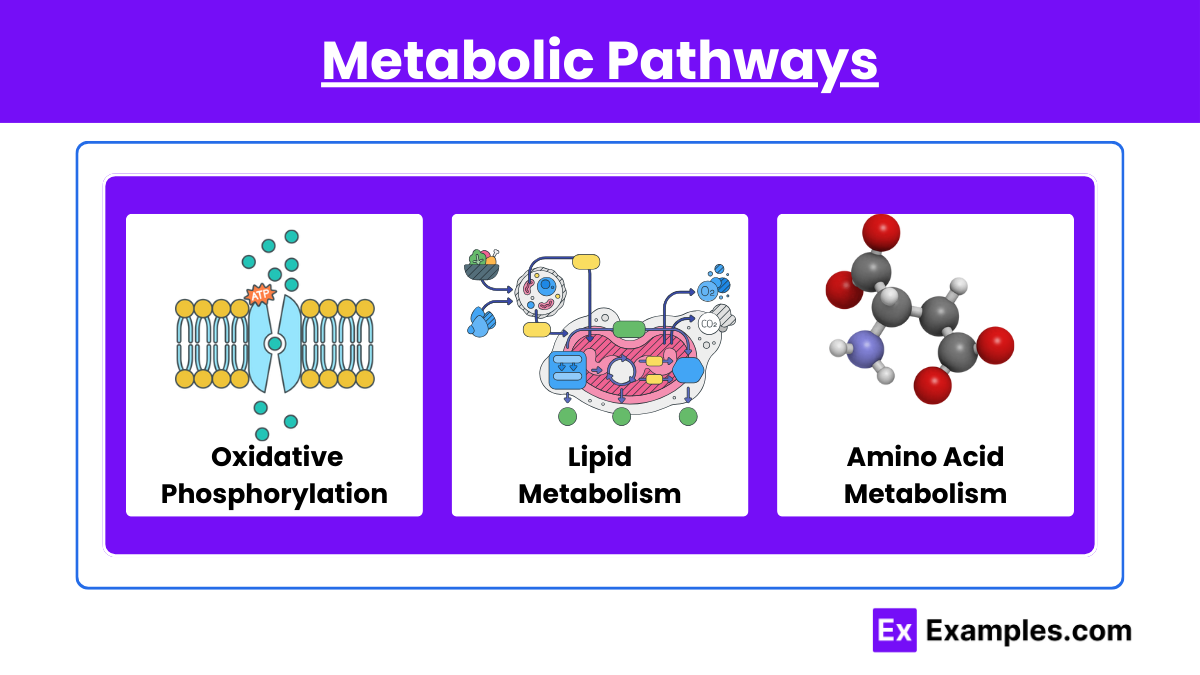
- Glycolysis, Citric Acid Cycle, and Oxidative Phosphorylation: Know each step’s reactants, products, and enzymes. Grasp the energy yield and the role of ATP, NADH, and FADH2. Understand how these pathways integrate and their regulation mechanisms.
- Gluconeogenesis and Glycogen Metabolism: Understand the reverse process of glycolysis in gluconeogenesis and the synthesis and breakdown of glycogen.
- Lipid Metabolism: Know beta-oxidation for fatty acid breakdown and how ketone bodies are formed and utilized, especially during fasting states.
- Amino Acid Metabolism: Grasp the concepts of transamination, oxidative deamination, and the urea cycle for nitrogen disposal.
3. Genetic Information Flow
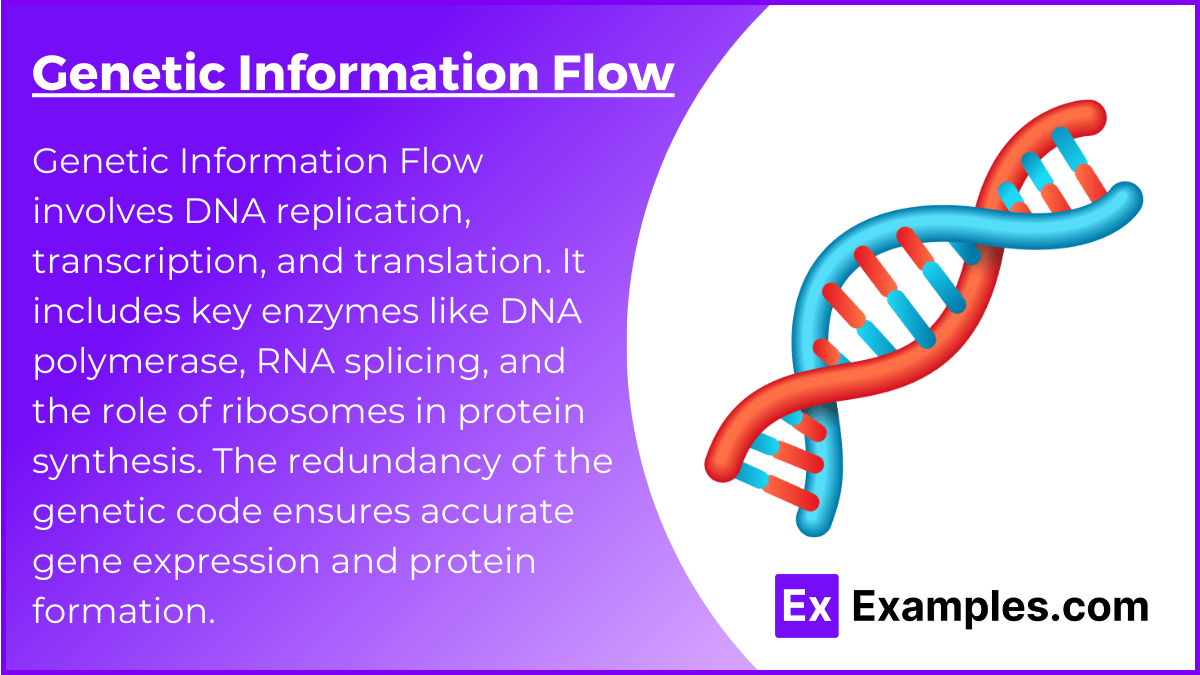
- DNA Replication and Repair: Cover the semi-conservative model, key enzymes (e.g., DNA polymerase, helicase, ligase), and mechanisms for proofreading and repair.
- Transcription and Post-Transcriptional Modifications: Understand transcription steps, RNA splicing, 5’ capping, and polyadenylation. Know the roles of different RNA types in gene expression regulation.
- Translation and Genetic Code: Know the process of translation, from initiation to termination, and the role of ribosomes. Understand codons and the concept of redundancy in the genetic code.
4. Regulation of Biological Systems
- Gene Expression Regulation: Understand prokaryotic vs. eukaryotic gene regulation (e.g., operons in prokaryotes, transcription factors in eukaryotes).
- Cell Signaling and Communication: Cover hormone signaling, signal transduction pathways, and the roles of secondary messengers like cAMP. Grasp concepts like autocrine, paracrine, and endocrine signaling.
- Homeostasis and Feedback Mechanisms: Know examples of negative and positive feedback mechanisms, particularly in endocrine pathways like the hypothalamic-pituitary axis.
Examples
Example 1: Enzyme Kinetics and Inhibition
A passage describes an experiment where a new enzyme’s activity is measured in the presence of various inhibitors. You’ll analyze data on reaction rates with competitive and noncompetitive inhibitors, calculate Km and Vmax values, and interpret enzyme-substrate affinity. Questions may involve predicting effects of enzyme mutations on reaction rates.
Example 2: Metabolism of Glucose in Cancer Cells
This passage presents data from a study investigating how cancer cells metabolize glucose differently from normal cells. You might explore glycolysis and the Warburg effect, comparing ATP yields and analyzing how inhibitors of glycolytic enzymes impact cancer cell viability. Questions may involve understanding altered metabolic pathways in cancer.
Example 3: Genetic Mutations and Protein Function
A scenario discusses a genetic disorder resulting from a single-point mutation affecting a protein’s function. You’ll examine how this mutation disrupts protein structure or enzyme activity and assess potential treatments. This passage could include data on enzyme kinetics, protein folding, or transcription and translation changes caused by the mutation.
Example 4: Cell Signaling Pathways and Hormonal Regulation
This passage explores how a specific hormone activates a signaling pathway that impacts gene expression. You may analyze data showing changes in secondary messenger levels and how these changes regulate metabolic activities. Questions may focus on understanding pathway components like G-proteins, receptor types, and cellular responses.
Example 5: Lipid Metabolism During Starvation
In this example, the passage discusses the body’s adaptation to starvation through lipid metabolism and ketone body formation. Data on blood glucose, free fatty acids, and ketone levels during prolonged fasting are provided. You’ll need to interpret the shift from glucose to fat utilization, understand beta-oxidation, and explain the role of ketones in energy production.
Practice Questions
Question 1:
Which of the following correctly describes the effect of a competitive inhibitor on an enzyme-catalyzed reaction?
A) It decreases both Km and Vmax.
B) It increases Km but does not change Vmax.
C) It decreases Vmax but does not change Km.
D) It increases both Km and Vmax.
Answer: B) It increases Km but does not change Vmax.
Explanation: Competitive inhibitors compete with the substrate for the active site, increasing Km by requiring more substrate to reach Vmax. Vmax remains unchanged as the maximum reaction rate can still be achieved with enough substrate.
Question 2:
During prolonged fasting, which metabolic shift occurs to provide energy for the brain?
A) Increased glucose utilization
B) Increased ketone body production
C) Decreased lipid metabolism
D) Increased protein synthesis
Answer: B) Increased ketone body production.
Explanation: Prolonged fasting triggers increased fat breakdown and ketone body production to supply energy, especially for the brain, as glucose stores are depleted and ketones become the primary fuel source.
Question 3:
A point mutation that changes a codon from UGU (cysteine) to UGA results in:
A) Silent mutation
B) Missense mutation
C) Frameshift mutation
D) Nonsense mutation
Answer: D) Nonsense mutation.
Explanation: UGA is a stop codon, resulting in a premature termination of translation. This nonsense mutation truncates the protein, often causing loss of function due to the incomplete polypeptide chain.